In order to support the development of personalized medicine in oncology, information about proteotype (identity, distribution, amount and organization of proteins, and how they relate to the genome) are anticipated to support improved clinical care for cancer patients. However, there is a strong need to standardize international efforts to deliver reliable and reproducible proteotype analysis. Towards this goal, international consortia have been formed to implement and standardize analytical methods.
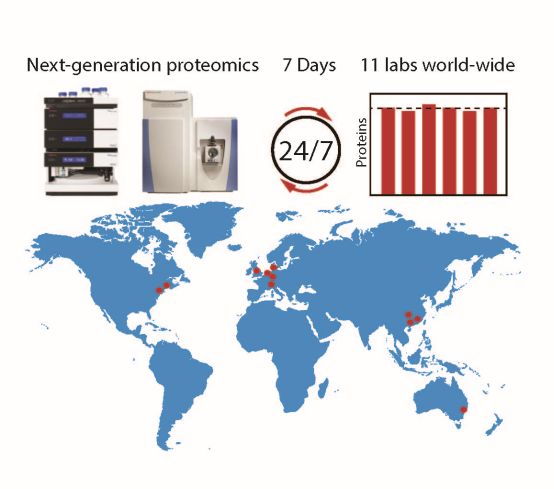
Sander Piersma and Connie Jimenez, of the OncoProteomics Laboratory at the VUmc, were part of an international effort to expand and integrate proteotype analysis in a clinical setting. Using harmonized mass spectrometry instrument platforms and standardized data acquisition procedures, the researchers demonstrated robust, sensitive, and reproducible data generation across eleven international sites on seven consecutive days in a 24/7 operational mode.
The novel next-generation proteomics workflow, based on data-independent acquisition mass spectrometry, revealed that coordinated and highly reproducible proteomics data acquisition is feasible from complex cancer samples. This work paves the way for precision medicine by proteome analysis of large clinical specimen cohorts across multiple labs.
Read the full article 'Standardization and harmonization of distributed multi-center proteotype analysis supporting precision medicine studies' in Nature Communications 2020.