Pancreatic ductal adenocarcinoma (PDAC) is a deadly form of cancer. Most patients with PDAC are diagnosed at an advanced stage and face a poor prognosis with limited treatment options. Thus, there is an urgent need for novel therapeutic strategies against PDAC.
A promising path to better treatments.
Unlike many other forms of cancer, PDAC has only a few 'driver' mutations – alterations in genes that propel the growth of cancer. This makes it more difficult to develop targeted treatments that home in on genetic changes.
However, cancer cells are not just defined by their genetic alterations; they also carry unique patterns of protein activities that can be exploited for therapy. A subset of one specific type of protein, known as kinases, are often highly active in cancer cells. Kinases play a crucial role in cellular processes such as division, survival, and metabolism. Activation of these proteins – which can be triggered by the addition of a phosphate group, a process known as phosphorylation – can cause kinases to drive the uncontrolled growth and division that characterizes cancer cells.
Consequently, targeting these hyperactive kinases has become a primary focus of many cancer treatments, with the aim to disrupt their signaling pathways and inhibit cancer progression.
‘Hyperactive’ proteins as cancer drivers
A research team from Cancer Center Amsterdam led by Prof. Connie Jiménez recently developed a comprehensive map of the activity of kinases in pancreatic ductal adenocarcinoma (PDAC) by applying phosphoproteomics. This technology can determine the phosphorylation status of thousands of proteins at once. The data were subsequently subjected to an integrative inferred kinase activity (INKA) analysis, an analysis pipeline developed by the OncoProteomics Laboratory, which can be accessed as a web-tool. In the present study, the research team studied nine different PDAC cell lines and identified more than 20,000 sites of phosphorylation activity on nearly 6,000 different proteins.
Triple drug combinations pack a punch
Based on the INKA data, several different kinase inhibitors were tested to block the activity of individual hyperactivated kinases. However, the single target approach only showed effect at high dosage levels that cannot be applied in the clinic mainly due to drug toxicity.
Next, the researchers tried treating the cancer cells with low doses of three different inhibitors, each targeting different activated kinases. They discovered this triple treatment was far more effective in curbing the growth of PDAC cells than single-drug treatments.
Towards a tailored therapeutic approach
Interestingly, the researchers found that the triple-drug approach was particularly more effective against a more aggressive subtype of PDAC known as the mesenchymal type.
Moreover, applying combinations of kinase inhibitors makes it harder for the cancer to acquire therapy resistance, and also supports the usage of these drugs at a low dose which is likely to give fewer side effects due to reduced toxicity.
Further research regarding the triple-drug cocktail is needed, including optimization for different PDAC subtypes followed by clinical trials in patients.
For more information, contact Prof. Connie Jiménez, or read the publication:
Vallés-Martí, A., et al (2023) Phosphoproteomics guides effective low-dose drug combinations against pancreatic ductal adenocarcinoma. Cell Reports 42, 112581. https://doi.org/10.1016/j.celrep.2023.112581
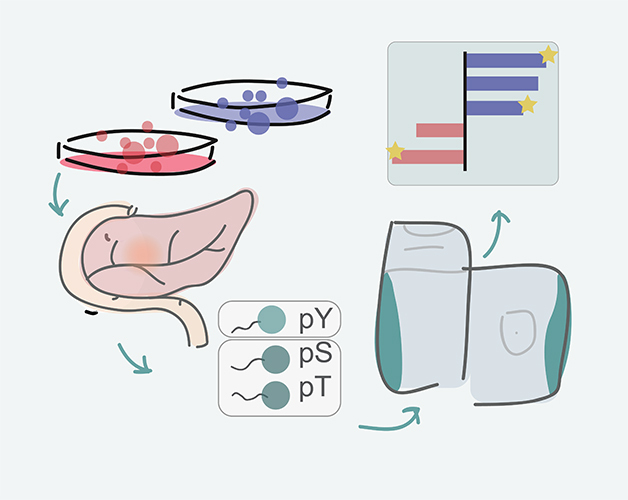
Visualisation of experimental outline: from tissue culture models of PDAC to identifying phosporylated amino acids in proteins to analysis pipeline and identification of promising drug targets.
People involved at Amsterdam UMC – Cancer Center Amsterdam
Andrea Vallés-Martí
Giulia Mantini
Paul Manoukian
Cynthia Waasdorp
Arantza Farina Sarasqueta
Richard R. de Goeij- de Haas
Alex A. Henneman
Sander R. Piersma
Thang V. Pham
Jaco C. Knol
Elisa Giovannetti
Maarten F. Bijlsma
Funding
This project was supported by the Dutch Cancer Society (KWF), European Union’s Horizon 2020 Research and Innovation Program under Marie Skłodowska-Curie grant agreement, and the Associazione Italiana per la Ricerca sul Cancro. Cancer Center Amsterdam and Netherlands Organization for Scientific Research (NWO) are acknowledged for support of the mass spectrometry infrastructure and Surfsara for computing infrastructure.
Text by Laura Roy.
This article was created for Cancer Center Amsterdam.
© 2023 New Haven Biosciences Consulting – All rights reserved.