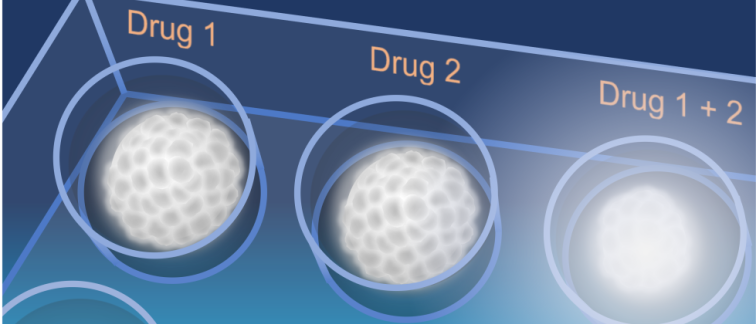
Glioblastoma is the most common primary brain cancer and one of the most lethal human
cancers. To improve patient outcomes, new treatments are urgently needed. To speed up the identification of new potential treatments against glioblastoma, the team of Bart Westerman has developed an artificial intelligence-powered imaging method to assess longitudinal cell growth after applying drug combinations to tumors cells.
Glioblastoma is a devasting disease with highly therapy-resistant tumor cells infiltrating healthy brain tissue. The blood-brain barrier, a natural protective ‘shield’ that protects the brain from toxic molecules in the blood stream, prevents many anti-cancer drugs from passing through and reaching glioblastoma tumors.
Applying a 3D culture model of glioblastoma tumors known as ‘neurospheres’, Bart Westerman and his team captured microscopic images of single neurospheres that had been exposed to various anti-cancer drug combinations over a period of 18 days. The captured images were then used to train a ‘Convolutional Neural Network’ (CNN) – a type of artificial intelligence that decomposes image data to recognize patterns. The CNN transformed image information was then trained to recognize patterns based on cell-viability tests, performed independently at the end of the experiments.
“The exciting thing about this research project is the innovative use of CNNs to convert microscopic images of glioblastoma cultures into data that explains the biochemistry that takes place in the tumor cells in a real time fashion,” says Anna Giczewska, PhD student and first author of the study which was published in Neuro-Oncology Advances.
From cell culture end-point to dynamic assessment of cell viability
The usual way of measuring the effectiveness of candidate drugs against tumor cells in cultures is to assess cell vitality as an endpoint of a growth experiment, frequently by lysing cells to release their intracellular ATP level that can be assessed by a chemiluminescent assay. However, this misses out on crucial information that could have been obtained on earlier time points since cells evidently need to be killed to measure their ATP levels.
“It is just as important - maybe more so - to capture the picture of how drugs might be influencing the growth of cells over the course of the experiment, not just checking how many cells are alive or dead at a certain time point. Although this is a proof of concept at this stage, it is an example of AI-based assessment of a biological phenotype as a function of biological interactions that take place into cells. The approach is likely to be valuable since continuous phenotype monitoring becomes possible which can also reveal new biological insights and show us how to move on experimentally.” says Bart Westerman, Associate Professor at Amsterdam UMC.
The team’s hypothesized that AI can be used to analyze the longitudinal imaging of neurosphere cell aggregates exposed to various drugs to uncover (often subtle) dynamics that change over time.
“Therefore, we segmented the images into smaller sections and processed them through a CNN decoder. We monitored 3D cell cultures over a period of three weeks in real-time and found, as proposed, that the appearance of cell aggregates can be linked to cellular ATP levels, reflective of the biochemistry that is taking place in the cells as a result of various drug combinations,” says Bart Westerman.
A new method to identify promising anti-cancer drug combinations
The team’s results showcase the value of image to data conversion and the power of AI to discover what escapes other means of detection. The method can be used to guide and refine further investigations to discover more effective drug combinations.
Anna Giczewska: “We demonstrated that longitudinal drug effects can be estimated from images using a CNN neural network strategy and we were able to assess drug interactions within a time frame of up to three weeks, which is unpreceded in this field”
The newly developed method will be put to work to screen and identify new drug combinations that can achieve higher efficacy against glioblastoma, including the ability to cross the blood-brain barrier.
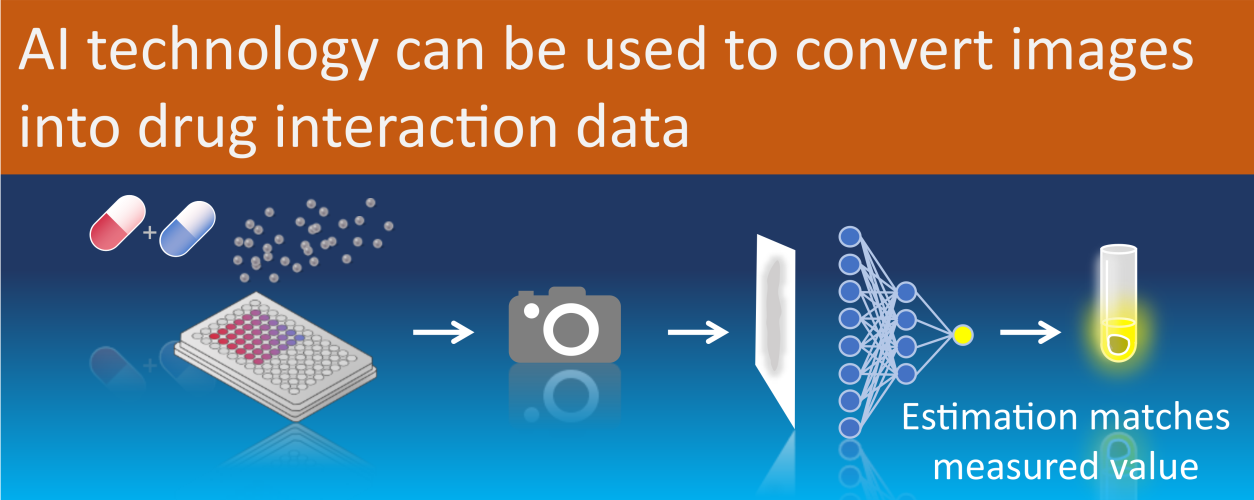
Visual representation of the workflow to assess cell viability using Convolutional Neural Networks.
For more information, contact Dr. Bart Westerman, or read the publication:
Anna Giczewska, A., et al. (2023) Longitudinal drug synergy assessment using convolutional neural network image-decoding of glioblastoma single-spheroid cultures. Neuro-Oncology Advances, vdad134, https://doi.org/10.1093/noajnl/vdad134.
Funding
Experimental work and wages were provided through funding by Cancer Center Amsterdam, Dutch Cancer Society Grant KWF-11038, Brain Tumour Charity Grant 488097, Health Holland AI Impact, and an APCA-PoC grant awarded by Amsterdam Innovation Exchange. The Tecan D300E Drug Printer was purchased with support from the Maurits en Anna de Kock Foundation. Furthermore, the companies NTRC Therapeutics B.V. and IOTA Pharmaceuticals contributed financially to this project.
Researchers involved at Cancer Center Amsterdam
Anna Giczewska
Krzysztof Pastuszak
Megan Houweling
Kulsoom Abdul
Noa Faaij
Laurine Wedekind
David Noske
Thomas Wurdinger
Anna Supernat
Bart Westerman
Text by Bart Westerman & New Haven Biosciences Consulting.
This article was created for Cancer Center Amsterdam.
Follow Cancer Center Amsterdam on LinkedIn & Twitter / X.
© 2023 New Haven Biosciences Consulting– All rights reserved.