With the increasing number of genetic diagnostic tests performed an increasing number of variants is identified of which the clinical significance is unknown. A functional test to assess pathogenicity can help solving these cases. Genes encoding proteins involved in chromatin remodeling, an important mechanism of regulation of gene expression, play a significant role in syndromal intellectual disability. Pathogenic variants in such genes result in an abnormal DNA methylation pattern throughout the genome which is highly specific for each protein or protein complex. This so called methylation signature can serve as a diagnostic marker for these syndromes and can help classify variants of unknown significance in these genes.
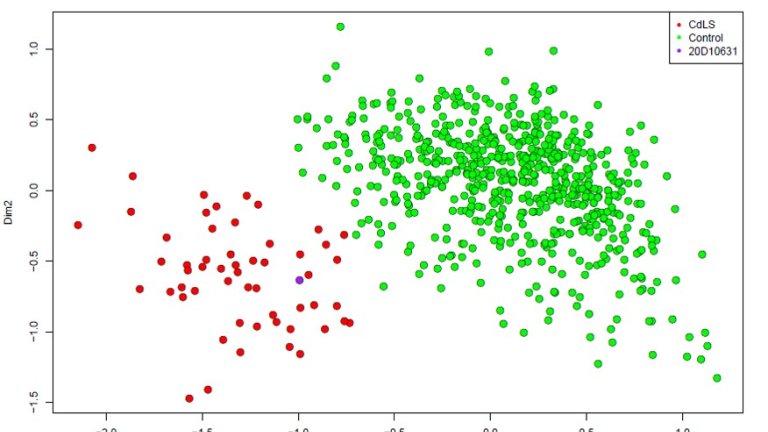
They have implemented a DNA methylation based test in diagnostics in collaboration with London Health Science Center in Canada. With the current version of the test (v2) they were able to diagnose 45 syndromes based on the methylation profile. A syndrome specific methylation signature in a patient with a variant of unknown significance will confirm the diagnosis and establish pathogenicity of the variant. In addition, also for patients without a molecular diagnosis EpiSign can be of use. The continuous identification of new signatures, adding more syndromes to the EpiSign panel will increase the diagnostic power and yield of this test. More than 20 new conditions will be added to the next EpiSign version (v3).
This project has generated the following publications:
A genome-wide DNA methylation signature for SETD1B-related syndrome
Krzyzewska IM et al. Clin Epigenetics. 2019 Nov 4;11(1):156
Evaluation of DNA Methylation Episignatures for Diagnosis and Phenotype Correlations in 42 Mendelian Neurodevelopmental Disorders
Aref-Eshghi E et al. Am J Hum Genet. 2020 Mar 5;106(3):356-370.
Clinical Epigenomics: Genome-Wide DNA Methylation Analysis for the Diagnosis of Mendelian Disorders
Sadikovic B et al. Genet Med. 2021 Feb 5.
Amsterdam UMC researchers involved in this project:
Marielle Alders, Marcel Mannens, Peter Henneman, Andrea Venema, Mieke van Haelst, Izabela Krzyzewska